Recently, I’ve been exploring species distribution modeling in R. My goal is to find new locations in Hawaii that are likely to yield C. elegans nematodes. Over the last 5 years, the Andersen Lab has collected over 5,000 substrates from five Hawaiian Islands and attempted to isolate C. elegans nematodes from all of them. So far, we’ve only been able to isolate C. elegans from ~5% of those collections.
We’ve noticed that the C. elegans positive samples mostly come from fruit substrates at high elevation in moderate moisture areas but we don’t know for certain what environmental parameters, if any, are important for limiting the distribution of C. elegans on the islands.
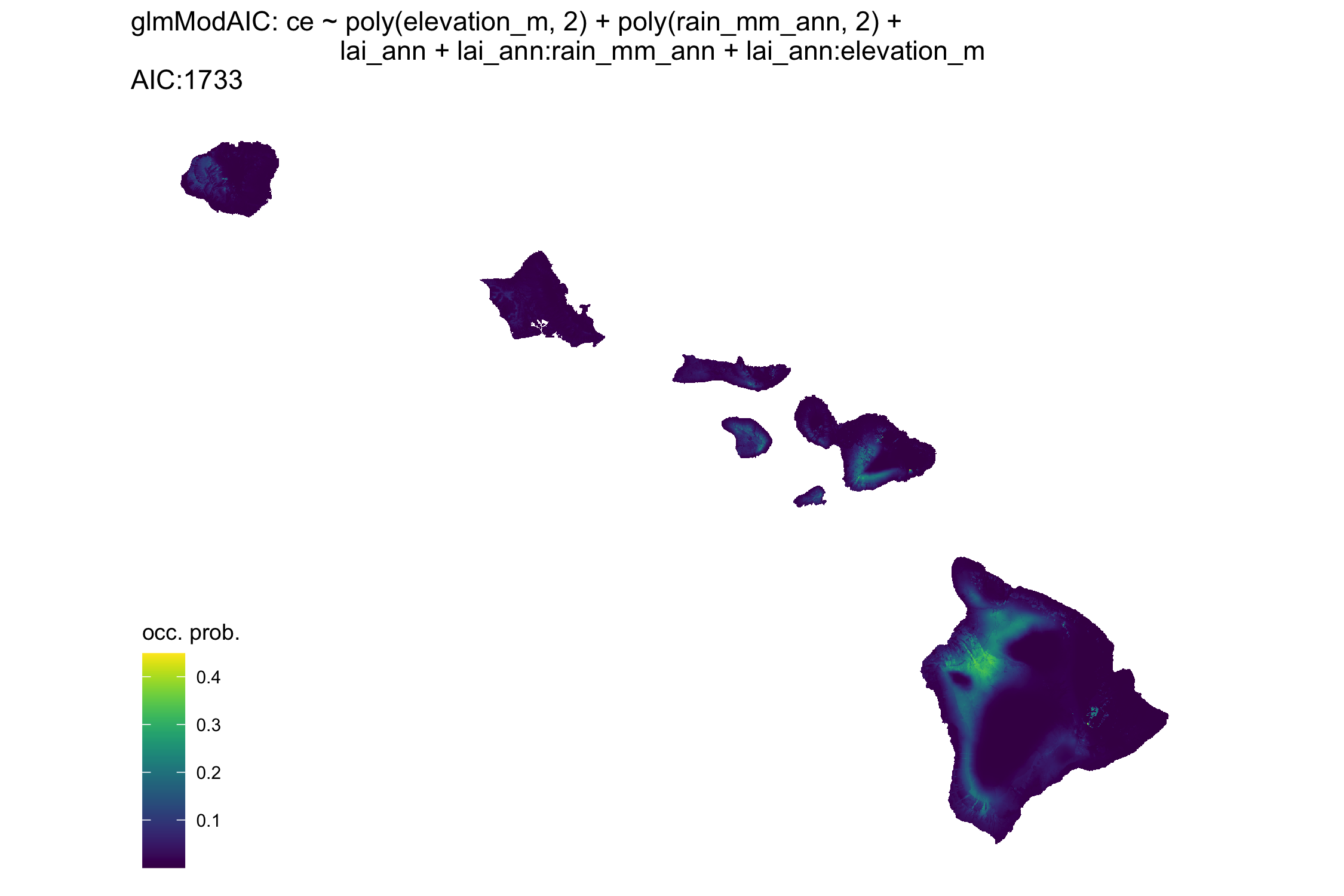
Here I’ve used spatial data from our collections and paired it with environmental data from raster files to predict the probability of C. elegans occurrence based on environmental factors. So far, I’ve built generalized linear models (glms) to predict presence/absence based on, elevation, annual leaf area coverage, and annual rainfall. I’ve also used tools to avoid collinearity and multi-collinearity among environmental predictors to reduce the complexity of the glms.
The plot above is an example of the best fit model according to AIC. Interestingly, there are locations we’ve never sampled before that have a high probability of occurrence - Southeast Maui here we come!